Wortmannin
|
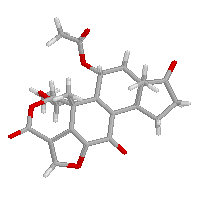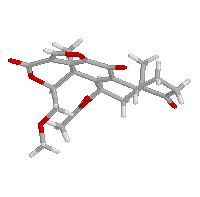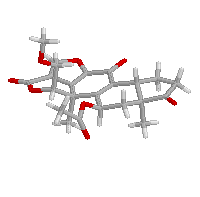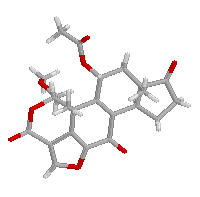 This structer was produced by RasMol, and stucked by GifBuilder (Yves Piguet, 1997) |
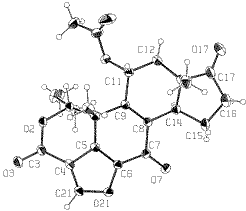 ORTEP-III (Burnett, 1996) drawing |
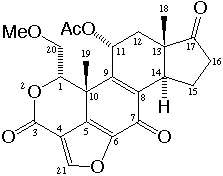
Wortmannin is isolated from Penicillium wortmanni and is a potent inhibitor
of phosphatidylinositol 3-kinase (PI 3-kinase). The electrophilic 21-site
makes a covalent linkage with the enzyme and the inhibition of PI 3-kinase
is irreversible. PI 3-kinase mediates several intracellular signaling
pathways, and wortmannin has been focused as a probe to understand the
structural requirements necessary for PI 3-kinase inhibition. Two types
of structure has been reported for wortmannin; one is orthorhombic crystal
and space group P2121212, the other is a 21-methylated analogue and
monoclinic, P21. This crystal is a polymorphic with monoclinic C2.
When the minimum van der Waals radii for carbon, oxygen and hydrogen atoms are considered approximately as 1.5, 1.7 and 1.2 Ang respectively, a short contact is found at C21...O21. The distance between H21 and O21 atoms is about 0.5 Ang below the sum of the van der Waals radii of hydrogen and oxygen (=2.7 Ang. and it could be defined as the C-H...O hydrogen bond. Paper: Polymorphism and C-H...O interactions of Wortmannin, a Phosphatidylinositol 3-Kinase Inhibitor Mitsunobu DOI, Toshimasa ISHIDA, Akira TERASHIMA, Taizo TANIGUCHI, Masahiro SASAKI and Chikako TANAKA Analytical Sciences (1998) 14, 1191-2. |
X-Ray Data Summary
| formula | C9 H10 F N O4 + H2O | ||
| weight | 233.20 | ||
| symmetry | monoclinic | ||
| space group | P21/c | ||
| Cell | Crystal | ||
| a (Ang) | 10.626(3) | description | needle |
| b (Ang) | 7.482(3) | colour | Colorless |
| c (Ang) | 13.065(4) | size (mm) | 0.5x0.09x0.09 |
| alpha (deg) | 90.0 | Dx (g/ml) | 1.553 |
| beta (deg) | 106.23(2) | F(000) | 488 |
| gamma (deg) | 90.0 | mu(CuKa) | 1.206 |
| V (Ang^3) | 997.4(6) | ||
| Z | 4 | ||
| Refinement | Diffrn measurement | ||
| Flack | device type | Rigaku AFC5R/RU-200 | |
| parameters | 147 | decay | 0.7 % |
| restraints | 0 | index limit | -12-h-12,-8-k-0,-15-L-15 |
| R_factor_all | 0.0592 | theta (deg.) | 4.33 - 65.58 |
| R_factor_gt | 0.0453 | total reflections | 3239 include Friedel pairs |
| wR_factor_ref | 0.1321 | reflections(obs) | 1393 .gt. 2sigma(I) |
| wR_factor_gt | 0.1186 | Structure | |
| shift/su_max | lt.0.001 | solution | SHELXS-97 |
| shift/su_mean | lt.0.001 | refinement | SHELXL-97 |
Get PDB coordinates
Get CIF text
Get SHELXL INS file
 Back to the structure index
Back to the structure index
Date: Jly.1998