KNI-272
|
Paper |
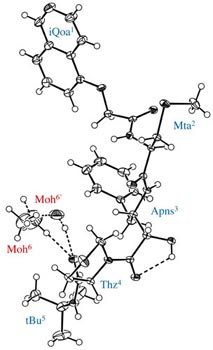
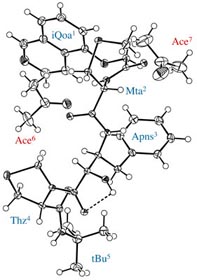
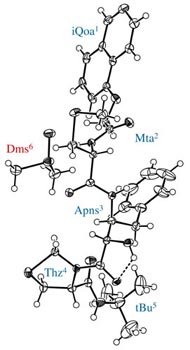
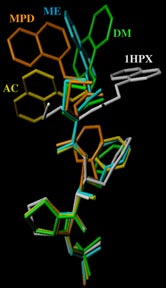
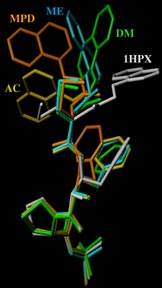
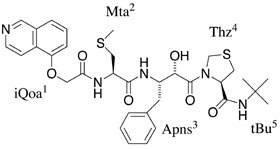
Rigid backbone moiety of KNI-272, a highly selective HIV protease inhibitor:
Methanol, acetone and dimethylsufoxide solvated forms of 3-[3-benzyl-2-hydroxy-9-(isoquinolin-5-yloxy)-6-methylsulfanylmethyl-5,8-dioxo-4,7-diazanonanoyl]-N-tert-butyl-1,3-thiazolidine-4-carboxamide. Mitsunobu Doi, Tooru Kimura, Toshimasa Ishida & Yoshiaki Kiso Acta Cryst. B60 (4), 433-437 (2004). Copyright © International Union of Crystallography |