1.1 Å resolution data
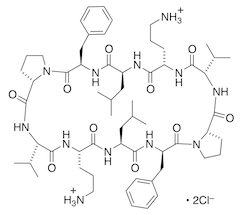
GS: cyclo(-Val-Orn-Leu-D-Phe-Pro-)2.2HCl.2HOH (+α HOH)
Space Group Orthorhombic, P21 21 2 _cell_length_a 34.988(1) _cell_length_b 23.381(1) _cell_length_c 18.8662(5) _cell_volume 15433.4(12) _cell_formula_units_Z 8 _diffrn_reflns_number 28227 _reflns_number_total 11453 _reflns_number_gt 9329 _diffrn_reflns_av_R_equivalents 0.0462 _diffrn_measurement_device_type 'Rigaku XtaLAB P200K' _refine_ls_R_factor_all 0.1793 _refine_ls_R_factor_gt 0.1650 _refine_ls_wR_factor_ref 0.4081 _refine_ls_wR_factor_gt 0.4007 Reference: Asano A & Doi M (2019) X-Ray Structure Analysis Online, 35, 1-2.
CCDC 1870209
PDB coordinates: GS2HCl.pdb
Map (SHELXL List 6) for coot: gsmap.fcf.zip (82 kB, right click and download)
INTENSITY STATISTICS Resolution #Data #Theory %Complete Redundancy Mean I Mean I/s R(int) R(sigma) Inf - 2.75 494 497 99.4 4.70 43.7 28.63 0.0362 0.0370 2.75 - 2.15 485 485 100.0 5.18 12.8 28.08 0.0475 0.0341 2.15 - 1.85 522 522 100.0 5.17 7.9 24.60 0.0601 0.0387 1.85 - 1.65 586 586 100.0 4.88 3.9 20.08 0.0788 0.0449 1.65 - 1.50 657 658 99.8 4.63 2.3 15.77 0.0896 0.0560 1.50 - 1.40 594 594 100.0 4.36 1.3 11.25 0.1166 0.0799 1.40 - 1.30 794 794 100.0 4.08 1.1 9.47 0.1217 0.0960 1.30 - 1.25 491 491 100.0 3.84 0.9 7.42 0.1608 0.1200 1.25 - 1.20 579 581 99.7 3.64 0.8 6.51 0.1725 0.1341 1.20 - 1.15 692 692 100.0 3.47 0.7 5.83 0.1886 0.1586 1.15 - 1.10 810 810 100.0 3.19 0.6 5.05 0.1954 0.1778 1.10 - 1.05 960 961 99.9 2.94 0.4 3.35 0.2610 0.2844 1.05 - 1.00 1173 1186 98.9 2.58 0.2 2.05 0.4074 0.4950 1.00 - 0.98 456 594 76.8 1.58 0.1 1.21 0.6194 0.9015 ------------------------------------------------------------------------------ Inf - 0.98 9293 9451 98.3 3.71 4.3 10.61 0.0565 0.0519
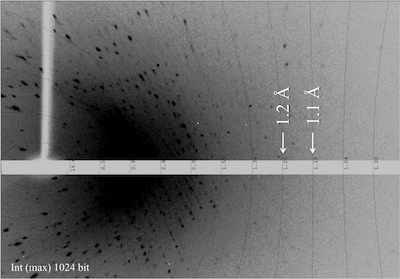
Diffraction image. Spots are observed over 1.1 Å, but weak..