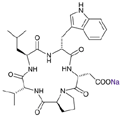
cyclo(-D-Trp-D-Asp--Pro-D-Val-Leu-).Na+, an endothelin inhibitor
Date: Oct.2000
|
BQ123 sodium salt cyclo(-D-Trp-D-Asp--Pro-D-Val-Leu-).Na+, an endothelin inhibitor Date: Oct.2000 |
 |
|
To display the left figure in your brawser, a tool, Chime, is necessary. Click buttons to show different view, and molecule will be rotated by your mouse. Shift and click = zoom; Option and click = translate |
|
Endothelin (ET) is the endogenous vasoconstriction peptide with 21
amino acid residues (Yanagisawa et al., Nature, 1988). Since its
activity is linked to the regulation of blood presser, this peptide has
been targeted to develop the antagonists expecting the therapeutic
applications for hypertension. Two cyclic peptapeptides of BE18257A and
B were isolated from Streptomyces misakiensis (Kojiri et al., J.
Antibiotics, 1991), which antagonized the ET-1 induced vasoconstriction
by binding to the endothelin A (ETA) receptor subtype. The more potent
ET-1 antagonists, BQ123, have been surveyed from these leading peptides
(Ihara et al., Life Sci., 1992). In addition to the screening of ET
antagonists, the structural studies have been performed for the
characterization of ET (Hewage et al., J. Pept. Sci., 1997), and the
helical property of the C-terminal region was focused (Okasa et al.,
Biochem. Biophys. Res. Commun., 1991; Van der Walle et al., J. Pharm.
Pharmacol., 1998). Such structural feature was also confirmed in the
crystal structure (Janes et al., Nature Struct. Biol.. et al. 1997).
The recent NMR study suggested that the structures of these antagonists
were partially similar to that of ET-1 (Coles et al., J. Med. Chem.,
1993; Peishoff et al., 1995). These structural studies for both
intrinsic substrates and antagonists are important to characterize the
structure-specificity of ETA receptor.
|
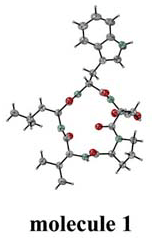
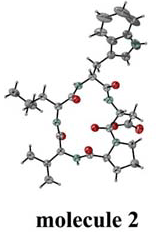
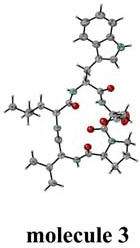
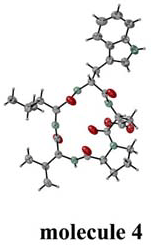 Fig.1. Four independent molecules. |
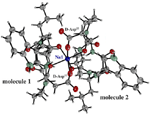
 Fig.2. Superimposition of four molecules. |
Formula 4(C31 H41 N6 O7 Na) Solvent(s) 8(H2 O),5.63(C3 H7 O H) Mr 3013.12 Crystal system orthorhombic Space group P212121 Radiation source SPring-8* a (A) 25.3930(4) Wavelength (A) 0.836 b (A) 34.7193(3) NREF mean. 83268 c (A) 25.7201(4) NREF 22865 Volume (A3) 22675.5(5) Resolution (A) 0.80 Z 4 Structure sol. LODEM Temperature (K) 100(2) Refinement SHELXL-97 Description block Npara 1904 Size (mm3) 0.08x0.04x0.04 R1 0.0576 Dx (g cm-1) 1.047 wR 0.1642 F(000) 6461 (shift/d)max 0.656 m (mm-1) 1.350 Drmax/min (e A3) 0.563/-0.263*Data were measured on a synchrotron, SPring-8/BL24XU-A, with the approval of Hyogo prefecture and Japan Synchrotron Radiation Research Institute (Approval No. C99A24XU-005N).
 Six coordinates: peptide:Na(2:1)
complexes - JPEG 60KB
Six coordinates: peptide:Na(2:1)
complexes - JPEG 60KB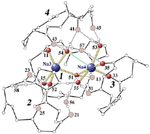 Five coordinates: Trigonal
and square pyramids - JPEG 56KB
Five coordinates: Trigonal
and square pyramids - JPEG 56KB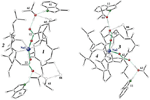 Electron relation from indol to sodium - JPEG 52KB
Electron relation from indol to sodium - JPEG 52KB
Na_1 - Distance Angles
O_22 2.2635 (0.0034)
O_12 2.2804 (0.0036) 168.77 (0.14)
OD1_12 2.3749 (0.0038) 101.17 (0.13) 81.82 (0.13)
OD1_22 2.3747 (0.0037) 79.79 (0.12) 111.26 (0.13) 87.78 (0.14)
O_24 2.4588 (0.0036) 81.70 (0.12) 87.13 (0.13) 99.93 (0.13) 161.01 (0.14)
O_14 2.4707 (0.0035) 95.23 (0.12) 81.89 (0.12) 163.58 (0.13) 96.17 (0.13) 81.34 (0.12)
Na_1 - O_22 O_12 OD1_12 OD1_22 O_24
Na_2 - Distance Angles
O_32 2.2470 (0.0035)
O_42 2.2881 (0.0035) 175.50 (0.15)
OD1_42 2.3377 (0.0039) 101.21 (0.14) 80.30 (0.13)
OD1_32 2.3874 (0.0046) 79.35 (0.14) 105.11 (0.14) 81.42 (0.16)
O_44 2.5021 (0.0035) 91.93 (0.13) 86.60 (0.12) 166.86 (0.14) 101.11 (0.15)
O_34 2.5191 (0.0038) 82.65 (0.12) 92.90 (0.13) 101.91 (0.15) 161.99 (0.14) 79.73 (0.12)
Na_2 - O_32 O_42 OD1_42 OD1_32 O_44
Na_3 - Distance Angles
O_11$3 2.2773 (0.0036)
O1_51 2.2919 (0.0036) 109.36 (0.14)
O1_54 2.3301 (0.0043) 88.67 (0.15) 88.67 (0.15)
O1_52 2.3404 (0.0038) 147.22 (0.15) 102.21 (0.14) 100.89 (0.14)
O_15$3 2.3843 (0.0039) 87.47 (0.13) 91.30 (0.13) 175.89 (0.14) 83.13 (0.13)
Na_3 - O_11$3 O1_51 O1_54 O1_52
Na_4 - Distance Angles
O1_55 2.2434 (0.0043)
O_33$4 2.3545 (0.0034) 119.56 (0.19)
O_35$4 2.3730 (0.0036) 85.48 (0.15) 103.46 (0.13)
O1_53 2.3758 (0.0041) 154.23 (0.19) 85.55 (0.13) 82.75 (0.14)
O1_51 2.3831 (0.0039) 81.50 (0.16) 83.47 (0.13) 166.98 (0.14) 109.03 (0.14)
O1_54 2.8472 (0.0042) 80.72 (0.16) 148.51 (0.13) 101.87 (0.12) 79.39 (0.12) 75.70 (0.12)
Na_4 - O1_55 O_33$4 O_35$4 O1_53 O1_51
Operators for generating equivalent atoms:
$1 -x+1/2, -y+2, z-1/2
$2 x-1/2, -y+3/2, -z+1
$3 -x+1/2, -y+2, z+1/2
$4 x+1/2, -y+3/2, -z+1