Valinyl bolaform amide
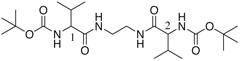 N,N'-Bis(t-buoxycarbonyl-valinyl)-diaminoethane |
2: 1 = 2 = L-Val 3: 1 = 2 = D-Val 4: 1 = L-Val, 2 = D-Val 5: 2 + 3 (1 : 1) mixture |
|
Paper |
Mitsunobu Doi, Akiko Asano, Hiroyuki Yoshida, Mizue Inouguchi, Kazunori Iwanaga, Masahiro Sasaki, Yoshio Katsuya, Taizo Taniguchi and Daisuke Yamamoto Structure and property of self-assemble valinyl bolaform amides having different chirality. J. Peptide Res. 22, 1234-1234 (2005). |
||
|
|||
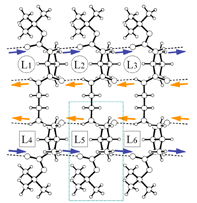  2 |
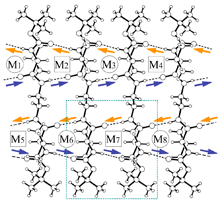  4 |
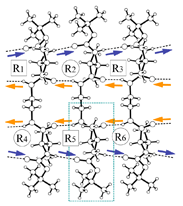  5 |
| Fig.1 β-sheet (crystal packing) and scanning electron microscopy | ||
| X-Ray summary for bolaform amides | |||
Cmin, mg/g (gelation) Tbot, K (melting) Formula Mr System Space group a b c β V Z T, K Dx F(0 0 0) Radiation Wavelength μ NREF (obs) Rint NREF (used) θmax R wR (Δ/σ)max Fraction θmax Δρmax Δρmin PDB coord. Packing x 40 mol |
2: L-peptide 9 397 C11H21N2O3 229.30 monoclinic C2 23.175(4) 5.0727(8) 16.310(2) 133.751(2) 1385.0(4) 4 200.0(2) 1.100 500 Mo Kα 0.7107 0.080 6081 0.0231 1686 27.1 0.0367 0.0953 <0.001 0.989 0.157 -0.139 lval.pdb L40.pdb |
4: meso-peptide 5 434 C22H42N4O6 458.60 monoclinic P21/n 9.4514(3) 17.8546(8) 17.7242(9) 94.190(4) 2983.0(2) 2 223(2) 1.021 1000 SPring-8/BL24XU 0.834 0.074 8773 0.0232 4533 29.0 0.0419 0.1144 0.001 0.926 0.222 -0.153 mval.pdb M40.pdb |
5: racemate 3 406 C11H21N2O3 229.30 tetragonal P-42(1)c 17.1628(8) 17.1628(8) 9.7150(9) 90.0 2861.7(3) 4 220(2) 1.064 500 Mo Kα 0.7107 0.077 32814 0.0256 1781 27.1 0.0390 0.1084 0.008 0.999 0.183 -0.110 dlval.pdb R40.pdb |
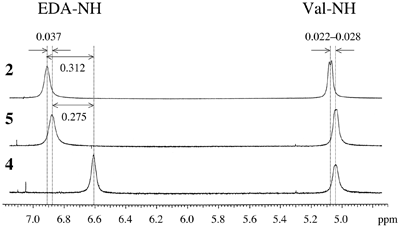 Fig.2 Chemical shifts of amide protons. Signal of amide proton EDA-NH of 4 is fairly shifted from those of 2 and 5, indicating the difference of H-bonds around EDA (Fig.1 center). |
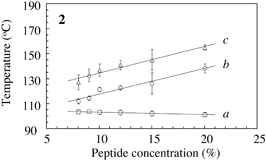
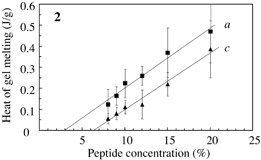
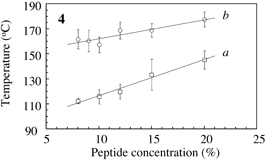
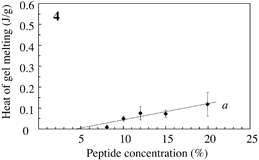
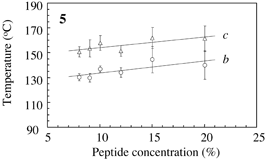
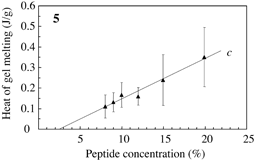
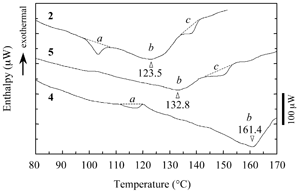 Fig.3 Typical DSC profile for decanol gels. Temperature of peak bottom (Tbot) could be an index for thermostability. Endthermal ΔH of peaks a, b and c are measured for peptide concentrations. |
 |  |
 |  |
 |  |
Back to the structure index
