 |
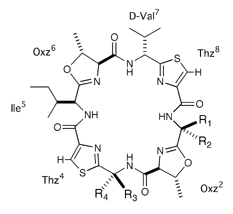
Asymmetirc ascidiacyclamide analogues
One amino acid was replaced to disturb C2-symmetry
Reference:
Akiko Asano, Katsuhiko Minoura, Takeshi Yamada, Atsushi Numata, Toshimasa
Ishida, Yoshio Katsuya, Yoshihiro Mezaki, Masahiro Sasaki, Taizo Taniguchi,
Masamichi Nakai, Hiroshi Hasegawa, Akira Terashima, and Mitsunobu Doi (2002)
J. Pept. Res. 60, 10-22.
Effect of asymmetric modification on the conformation of ascidiacyclamide analogues.
|
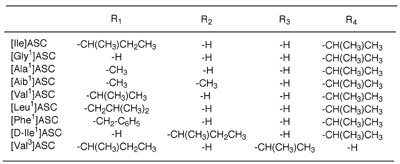 |
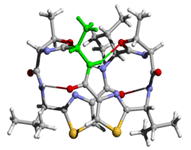
[Ala1]ASC (folded) |
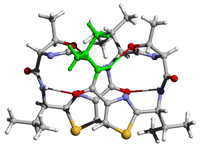
[Aib1]ASC (folded) |
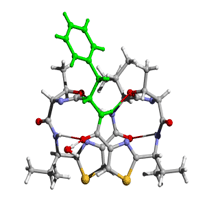
[Phe1]ASC (folded) |
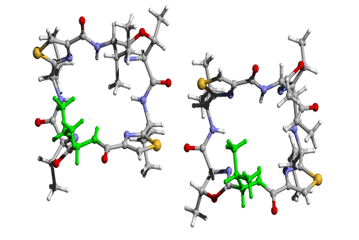
[Val1]ASC (square) |
|
These figures were produced from Rasmol and
rendered by POVray3 |
Table 1. Crystal and experimental data for ASC analogues
[Ala1]ASC [Phe1]ASC [Val1]ASC [Aib1]ASC
Formula C33H46N8O6S2 C39H50N8O6S2.H2O 2(C35H50N8O6S2) C34H48N8O6S2
Mr 714.9 809.1 1485.9 728.92
Crystal system monoclinic orthorhombic triclinic monoclinic
Space group P21 P212121 P1 P21
a, A 12.871(7) 8.4525(1) 10.6287(2) 12.9839(4)
b, A 10.159(5) 14.3982(2) 14.1562(6) 10.1151(3)
c, A 14.377(7) 33.8976(5) 14.3489(5) 14.4747(5)
a, deg 90 90 72.429(2) 90
b, deg 103.01(4) 90 83.660(2) 104.123(2)
g, deg 90 90 82.641(3) 90
V, A3 1831.6(16) 4125.4(1) 2035.4(1) 1843.55(10)
Z 2 4 2 2
T, K 240 240 100 100
Dx, g cm-3 1.301 1.303 1.212 1.313
F(000) 832 1720 792 776
X-ray source Cu Ka synchrotron synchrotron synchrotron
lambda, A 1.5418 0.834 0.835 0.835
m, mm-1 1.822 0.187 0.182 0.199
No. of reflx 4682 4855 7441 3947
No. of para 450 515 940 461
R 0.0593 0.0390 0.0667 0.0435
wR 0.1479 0.0973 0.1850 0.1236
Flack x -0.03(3) 0.54(8) 0.15(9) 0.04(10)
(D/s)max < 0.001 < 0.001 0.001 < 0.001
Drmax, e A3 0.498 0.362 1.018 0.392
Drmin, e A3 -0.275 -0.294 -0.480 -0.409
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Get PDB yes yes yes yes
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
The beam time of synchrotron, SPring-8/BL24XU-A, for this study was provided
by Hyogo Prefecture and the Japan Synchrotron Radiation Research Institute
(Approval No. C99A24XU-005N).
Analogues were further analyzed by NMR and CD spectroscopic methods. Summary
is listed in Table 2.
Table 2. Summary of structural properties and cytotoxic activities of ASC analogues
Form(a) Temp. Solvent dep(d) Conc. CD spectra(f) Cyto-
X-ray NMR(b) dep(c) DMSO acetone dep(e) MeCN TFE toxicity(g)
[Gly1]ASC F ++ +++ [+, 0] [++, ++] (-)
[Ala1]ASC F + ++ ++ + [+, 0] [++, ++] +-
[Aib1]ASC F ++ + ++ + [+, 0] [++, ++] -
[Val1]ASC S + ++ + + [++, -] [++, ++] +++
[Ile]ASC S(h) + ++ ++ + [+, -] [++, ++] ++
[Leu1]ASC S(i) S + ++ ++ + [+, +] [++, ++] (++)
[Phe1]ASC F F + ++ ++ ++ [+-, +] [++, ++] ++
[D-Ile1]ASC +++ ++ +++ ++ [++, 0] [++, ++] +
[Val3]ASC +++ +++ +++ + [+, -] [+, -] ++
a 'F' and 'S' represent folded and square forms, respectively.
b Molecular dynamics simulations are reported.
c Number of signs shows degree of temperature dependences (Dd/DT).
d Number of signs shows intensity of solvent effects.
e Number of signs shows effect of peptide concentration on chemical shifts.
f [q] values of 250-210 nm and 280-250 nm are indicated by left and right signs
in brackets, respectively.
g Number of signs shows degree of cytotoxic activity in P388 cells.
Activities taken from the previous report using L1012 cells are shown in parentheses.
h Crystal structures from various solvent systems were analyzed, and
all structures showed the square form .
i The structure is taken from the previous report.
Back





