Program:sortatom.exe.zip (Windows/32bit) /
sortatom.zip (OSX10.7/Intel i5)
Input eg: test1.res
Output eg1: test1a.res auto arrange
Output eg1: test1b.res from the strat atom
*Download from the right-click menu.
Input
test1.res :
The coorinates with SHELX format. The early structure of cyclic decapetpide Tyrocidine-B c[Val-Orn(OBzl)-Leu-DPhe-Pro-Trp-DPhe-Asn-Gln-Phe].
TITL Tyrocidine-B(Z) P212121 CELL 0.836 34.1331 9.5394 30.2385 90.000 90.000 90.000 ZERR 4.00 0.0068 0.0019 0.0060 0.000 0.000 0.000 LATT -1 SYMM 0.5-X,-Y,0.5+Z SYMM 0.5+X,0.5-Y,-Z SYMM -X,0.5+Y,0.5-Z SFAC C H N O UNIT 272 352 56 52 TEMP -173 C001 1 0.43620 0.55638 0.46053 11.000 0.05 99.00 C002 1 0.44997 0.77481 0.61016 11.000 0.05 91.17 C003 1 0.53326 1.09592 0.58496 11.000 0.05 90.35 C004 1 0.44117 0.65702 0.49400 11.000 0.05 88.21 C005 1 0.50497 0.98884 0.59736 11.000 0.05 86.49 C006 1 0.44501 0.90630 0.63609 11.000 0.05 83.58 C007 1 0.44795 0.63107 0.54113 11.000 0.05 81.95 ...
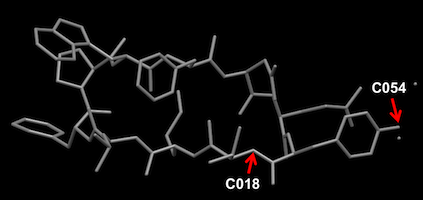
Mercury showing.
The return code of Windows is different from OSX. OSX file has CR, but this return code does not work for OSX programs. Please change CR to LF (Linux) or CR+LF (win).
Usage
SHELXS outputs test1.res with the peaks height order. WInGX or Mercury shows the molecular structure.
For numbering test1.res with the chemical structure, the peaks height order forces the huge patient.
1 2 3 4 5 6 7 8 9 10 c[Val-Orn(OBzl)-Leu-DPhe-Pro-Trp-DPhe-Asn-Gln-Phe] C001 ---> D-Phe7 C = C072 C002 ---> Gln9 N = N091 C003 ---> Phe10 Cα = C101 C004 ---> Asn8 N = N081 ...Numbring like the blue letters, and re-arranege the atoms along the chemical bonds.
プログラムもデータも同じフォルダーに保存します。

meibomatch.exe をダブルクリックすると、MS-DOSのコマンドプロンプトが現れます。

あとは必要なデータを入力します。入力は部分は赤色です。
自動並べ替え(分子構造の一番端を起点にした並べ替え)
SORT atoms array along structure
================================
Input RES/INS file: test1.res ← 入力
Output file: test1a.res ← 出力
>> Read 97 atoms
Need atom-list? [Y/N*]: [リターン]
:: SORTING from
[*Cr] THE ATOM HAVING THE LONGEST VECTOR FROM CENTER
[2] INPUT ATOM
Which ?: [リターン]
↑ これで2原子の組合せの中で最も距離が長い2原子を選び、
どちらか一方を並び替えの先頭にもってくる
>> SORT started at: C001
>> Remaining 2 atoms are NOT devided to any fragment.
These atoms have no fragment number.
↑ 結合を調べたところ、2つの原子がどのクラスターにも属さない
------- OK? :
>> SORT started at: C054 ← C054を結合のスタート原子に設定
Job finshed. OK?
結果1
test1a.res の座標はC054(上図Mercuryの表示を参照)から、結合可能原子が順に並ぶ。その選び方にルールはないので、原子のリスト順は化学的なナンバリングと必ずしも一致しない。
TITL Tyrocidine-B(Z) P212121 CELL 0.836 34.1331 9.5394 30.2385 90.000 90.000 90.000 ZERR 4.00 0.0068 0.0019 0.0060 0.000 0.000 0.000 LATT -1 SYMM 0.5-X,-Y,0.5+Z SYMM 0.5+X,0.5-Y,-Z SYMM -X,0.5+Y,0.5-Z SFAC C H N O UNIT 272 352 56 52 RESI 0 ↓ クラスタ番号 C054 1 0.60204 0.92606 0.77591 11.00 0.05000 1 C024 1 0.59653 0.94390 0.73219 11.00 0.05000 1 C025 1 0.59546 0.83680 0.70070 11.00 0.05000 1 C038 1 0.59319 1.08381 0.71896 11.00 0.05000 1 C045 1 0.58760 0.86173 0.65593 11.00 0.05000 1 C048 1 0.58644 1.10231 0.67361 11.00 0.05000 1 C015 1 0.58342 1.00083 0.64022 11.00 0.05000 1 C019 1 0.57614 1.04045 0.59036 11.00 0.05000 1 C003 1 0.53326 1.09592 0.58496 11.00 0.05000 1 C005 1 0.50497 0.98884 0.59736 11.00 0.05000 1 ....
基点を指定して並べ替え
SORT atoms array along structure
================================
Input RES/INS file: test1.res
Output file: test1b.res
>> Read 97 atoms
Need atom-list? [Y/N*]: [リターン]
:: SORTING from
[*Cr] THE ATOM HAVING THE LONGEST VECTOR FROM CENTER
[2] INPUT ATOM
Which ?: 2 ← 基点原子入力を選択
------- OK? :
:: ATOMS of FRAGMENT 1
C001 C004 C009 C039 C007 C017 C040 C013 C032 C027
C026 C002 C011 C022 C029 C047 C042 C044 C006 C031
C051 C014 C036 C064 C052 C028 C046 C075 C041 C049
C005 C034 C012 C073 C088 C055 C072 C003 C008 C062
C085 C074 C078 C081 C019 C021 C020 C023 C063 C066
C067 C089 C092 C015 C018 C035 C056 C080 C094 C045
C048 C010 C069 C079 C025 C038 C016 C033 C058 C077
C024 C030 C037 C065 C068 C043 C050 C070 C087 C054
C059 C053 C086 C071 C082 C057 C084 C091 C076 C060
C097 C090 C093 C096 C095
INPUT START ATOM ([Cr]=Atom having the LONGEST vector): C018
原子名は大文字小文字も区別される。もし、一致する原子名がないと自動選択に戻る。
>> SORT started at: C018
> This fragment has 95 atoms
C018 C021 C010 C003 C035 C016 C033 C005 C019 C030
C037 C065 C068 C028 C015 C059 C006 C034 C045 C048
C057 C084 C002 C046 C025 C038 C053 C060 C097 C013
C012 C024 C050 C096 C007 C011 C008 C054 C058 C086
C095 C004 C032 C020 C023 C069 C043 C091 C001 C022
C056 C090 C093 C009 C039 C031 C051 C066 C079 C017
C040 C062 C080 C077 C027 C026 C073 C063 C070 C087
C029 C047 C042 C044 C075 C085 C067 C071 C082 C014
C036 C064 C052 C088 C076 C041 C049 C055 C072 C074
C078 C081 C089 C092 C094
> REMAINING: 2 ATOMS
Job finshed. OK?
結果2
test1b.res の座標はC018(上図Mercuryの表示を参照:N(Val1))から、結合可能原子が順に並ぶ。その選び方にルールはないので、原子のリスト順は化学的なナンバリングと必ずしも一致しない。
TITL Tyrocidine-B(Z) P212121 CELL 0.836 34.1331 9.5394 30.2385 90.000 90.000 90.000 ZERR 4.00 0.0068 0.0019 0.0060 0.000 0.000 0.000 LATT -1 SYMM 0.5-X,-Y,0.5+Z SYMM 0.5+X,0.5-Y,-Z SYMM -X,0.5+Y,0.5-Z SFAC C H N O UNIT 272 352 56 52 RESI 0 C018 1 0.50674 1.07911 0.50833 11.00 0.05000 1 C021 1 0.52722 1.14772 0.53826 11.00 0.05000 1 C010 1 0.49905 1.12708 0.46133 11.00 0.05000 1 C003 1 0.53326 1.09592 0.58496 11.00 0.05000 1 C035 1 0.54418 1.26727 0.52441 11.00 0.05000 1 C016 1 0.46333 1.06097 0.44595 11.00 0.05000 1 C033 1 0.53524 1.09180 0.42778 11.00 0.05000 1 C005 1 0.50497 0.98884 0.59736 11.00 0.05000 1 C019 1 0.57614 1.04045 0.59036 11.00 0.05000 1 C030 1 0.43025 1.13344 0.43789 11.00 0.05000 1 C037 1 0.45863 0.91801 0.44269 11.00 0.05000 1 ....
□ □ □